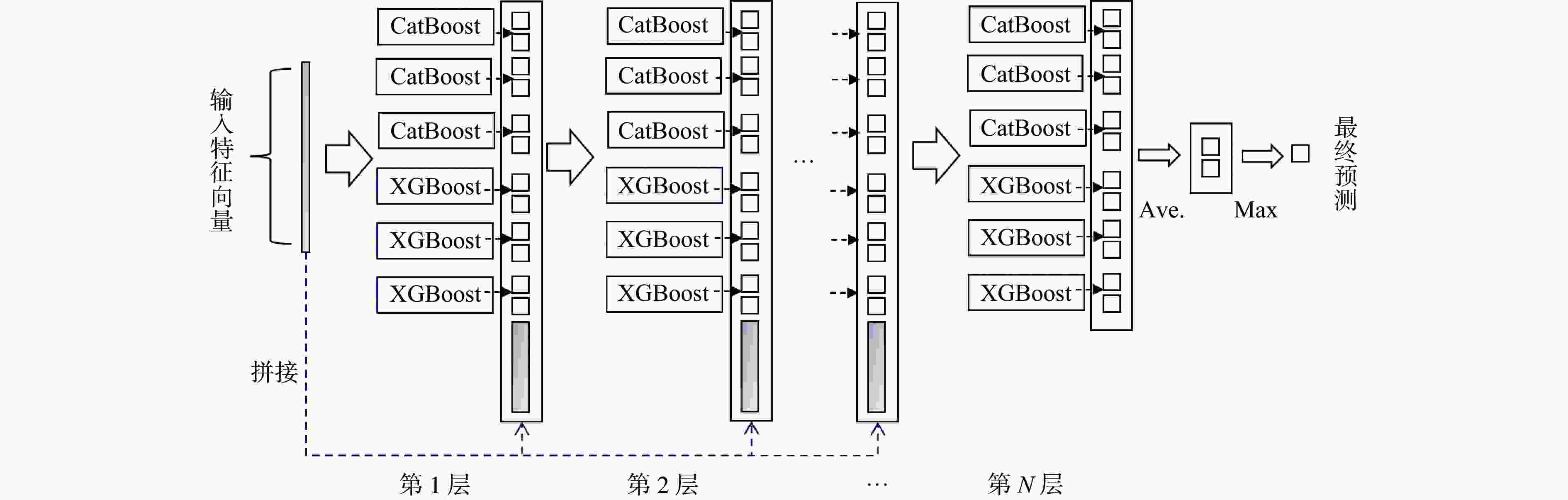
Predicting drug-target interactions based on dynamic multi-grained scanning
-
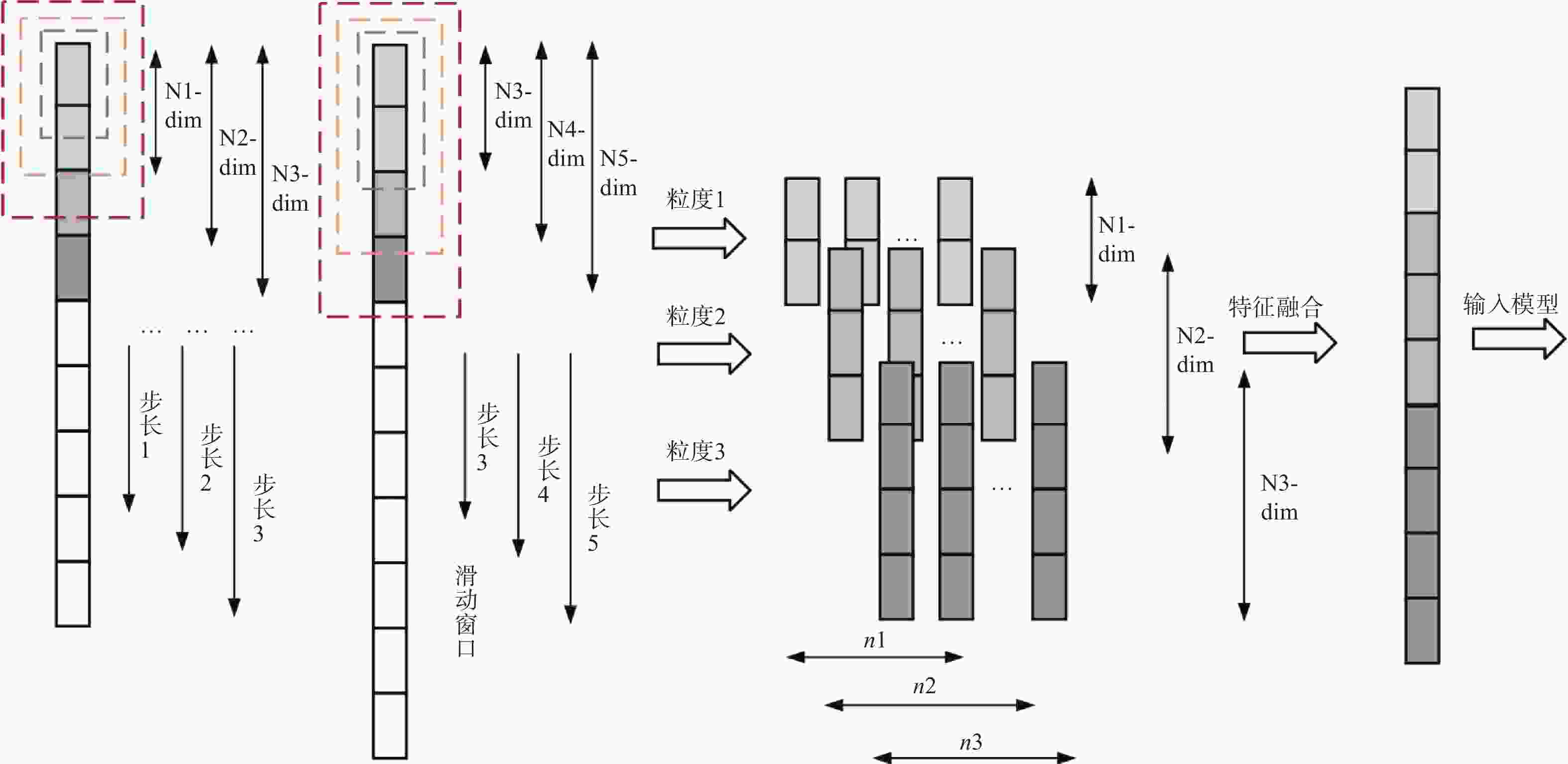
摘要: 针对传统机器学习模型在药物−靶点预测任务中由浅层模型结构和复杂数据特征导致分类表现不佳的问题,提出一种新预测模型DMS-DF。该模型基于深度森林算法,引入动态自适应多粒度扫描机制,并选择CatBoost和XGBoost作为级联森林基分类器。结果表明, DMS-DF模型在药物–靶点预测中表现优于同一数据集下的其他4个模型,为药物发现提供了新途径。Abstract: To address the poor classification performance of traditional machine learning models in the drug-target prediction, a problem caused by their shallow structure and complex data features, a novel prediction model DMS-DF was proposed. The model was based on the deep forest algorithm, the model incorporated a dynamic adaptive multi-granularity scanning mechanism. Furthermore, CatBoost and XGBoost were selected as cascade forest-based classifiers. It demonstrates that that the DMS-DF model outperforms the other four models in terms of drug-target prediction on the same dataset, providing a novel approach for drug discovery.
-
表 1 DMS-DF和其他方法的表现
Table 1. Performance of DMS-DF and baseline methods
模型 Sn Sp MCC AUC AUPR DMS-DF 0.9417 0.9317 0.8935 0.9847 0.9857 LGBMDF 0.9451 0.9471 0.8924 0.9844 0.9855 NEDTP 0.9194 0.9267 0.8462 0.9714 0.9690 SVM 0.8869 0.9286 0.8162 0.9668 0.9664 RF 0.9138 0.9348 0.8488 0.9784 0.9798 表 2 每种方法的性能比较
Table 2. Performance comparison under each method
模型 AUC AUPR 3XGBoost-3RF 0.9813 0.9834 3CatBoost-3RF 0.9796 0.9818 DMS-DF 0.9847 0.9857 -
[1] SCHOMBURG I, CHANG A, PLACZEK S, et al. BRENDA in 2013: integrated reactions, kinetic data, enzyme function data, improved disease classification: new options and contents in BRENDA[J] . Nucleic Acids Research, 2013, 41: D764 − D772. [2] LOTFI SHAHREZA M, GHADIRI N, MOUSAVI S R, et al. A review of network-based approaches to drug repositioning[J] . Briefings in Bioinformatics, 2018, 19(5): 878 − 892. doi: 10.1093/bib/bbx017 [3] KANEHISA M, FURUMICHI M, TANABE M, et al. KEGG: new perspectives on genomes, pathways, diseases and drugs[J] . Nucleic Acids Research, 2017, 45: D353 − D361. doi: 10.1093/nar/gkw1092 [4] LEE I, KEUM J, NAM H. DeepConv-DTI: prediction of drug-target interactions via deep learning with convolution on protein sequences[J] . PLoS Computational Biology, 2019, 15(6): e1007129. doi: 10.1371/journal.pcbi.1007129 [5] RU X Q, YE X C, SAKURAI T, et al. Current status and future prospects of drug–target interaction prediction[J] . Briefings in Functional Genomics, 2021, 20(5): 312 − 322. doi: 10.1093/bfgp/elab031 [6] LIU Y, WU M, MIAO C Y, et al. Neighborhood regularized logistic matrix factorization for drug-target interaction prediction[J] . PLoS Computational Biology, 2016, 12(2): e1004760. doi: 10.1371/journal.pcbi.1004760 [7] O'CONNELL M J, LOCK E F. Linked matrix factorization[J] . Biometrics, 2019, 75(2): 582 − 592. doi: 10.1111/biom.13010 [8] GIRYES R, SAPIRO G, BRONSTEIN A M. Deep neural networks with random Gaussian weights: a universal classification strategy?[J] . IEEE Transactions on Signal Processing, 2015, 64: 3444 − 3457. doi: 10.1109/TSP.2019.2961228 [9] BLEAKLEY K, YAMANISHI Y. Supervised prediction of drug-target interactions using bipartite local models[J] . Bioinformatics, 2009, 25(18): 2397 − 2403. doi: 10.1093/bioinformatics/btp433 [10] YAMANISHI Y, KOTERA M, KANEHISA M, et al. Drug-target interaction prediction from chemical, genomic and pharmacological data in an integrated framework[J] . Bioinformatics, 2010, 26(12): i246 − i254. doi: 10.1093/bioinformatics/btq176 [11] 白茹, 滕奇志, 杨晓敏, 等. 基于SVM和GA的药物与人血清白蛋白结合的预测[J] . 计算机工程与应用, 2009, 45(12): 226 − 228, 248. doi: 10.3778/j.issn.1002-8331.2009.12.072 [12] GÖNEN M. Predicting drug–target interactions from chemical and genomic kernels using Bayesian matrix factorization[J] . Bioinformatics, 2012, 28(18): 2304 − 2310. doi: 10.1093/bioinformatics/bts360 [13] YAN X, YOU Z H, WANG L, et al. DTIFS: a novel computational approach for predicting drug-target inter-actions from drug structure and protein sequence[C] //Proceedings of the 16th International Conference on Intelligent Computing Theories and Application. Bari: Springer, 2020: 371 − 383. [14] LIAN M J, DU W L, WANG X J, et al. Drug-target interaction prediction based on multi-similarity fusion and sparse dual-graph regularized matrix factorization[J] . IEEE Access, 2021, 9: 99718 − 99730. doi: 10.1109/ACCESS.2021.3096830 [15] 章新友, 王芝, 张春强, 等. 相似性算法在药物−靶点预测研究中的应用[J] . 中国新药杂志, 2024, 33(9): 885 − 894. doi: 10.3969/j.issn.1003-3734.2024.09.007 [16] WANG Y C, YANG Z X, WANG Y, et al. Computationally probing drug-protein interactions via support vector machine[J] . Letters in Drug Design & Discovery, 2010, 7(5): 370 − 378. [17] 刘文昌, 魏赟, 袁浩轩, 等. 基于SMOTE和gcForest的医疗小样本数据分类研究[J] . 物联网学报, 2023, 7(2): 76 − 87. doi: 10.11959/j.issn.2096-3750.2023.00337 [18] CHAWLA N V, BOWYER K W, HALL L O, et al. SMOTE: synthetic minority over-sampling technique[J] . Journal of Artificial Intelligence Research, 2002, 16: 321 − 357. doi: 10.1613/jair.953 [19] GOODFELLOW I, BENGIO Y, COURVILLE A. Deep learning[M] . Cambridge: The MIT Press, 2016: 216. [20] ZHOU Z H, FENG J. Deep forest: towards an alternative to deep neural networks[C] // Proceedings of the 26th International Joint Conference on Artificial Intelligence. Melbourne: ACM, 2017: 3553 − 3559. [21] PROKHORENKOVA L, GUSEV G, VOROBEV A, et al. CatBoost: unbiased boosting with categorical features[C] //Proceedings of the 32nd International Conference on Neural Information Processing Systems. Montréal: ACM, 2018: 6639 − 6649. [22] CHEN T Q, GUESTRIN C. XGBoost: a scalable tree boosting system[C] //Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining. San Francisco: ACM, 2016: 785 − 794. [23] CORTES C, VAPNIK V. Support-vector networks[J] . Machine Learning, 1995, 20(3): 273 − 297. [24] HO T K. Random decision forests[C] //Proceedings of 3rd International Conference on Document Analysis and Recognition. Montreal: IEEE, 1995: 278 − 282. [25] PENG Y, ZHAO S W, ZENG Z L, et al. LGBMDF: a cascade forest framework with LightGBM for predicting drug-target interactions[J] . Frontiers in Microbiology, 2022, 13: 1092467. doi: 10.3389/fmicb.2022.1092467 [26] AN Q, YU L. A heterogeneous network embedding framework for predicting similarity-based drug-target interactions[J] . Briefings in Bioinformatics, 2021, 22(6): 1 − 10. doi: 10.1093/bib/bbab275 -




 下载:
下载: